Authors:
(1) Arcangelo Massari, Research Centre for Open Scholarly Metadata, Department of Classical Philology and Italian Studies, University of Bologna, Bologna, Italy {arcangelo.massari@unibo.it};
(2) Fabio Mariani, Institute of Philosophy and Sciences of Art, Leuphana University, Lüneburg, Germany {fabio.mariani@leuphana.de};
(3) Ivan Heibi, Research Centre for Open Scholarly Metadata, Department of Classical Philology and Italian Studies, University of Bologna, Bologna, Italy and Digital Humanities Advanced Research Centre (/DH.arc), Department of Classical Philology and Italian Studies, University of Bologna, Bologna, Italy {ivan.heibi2@unibo.it};
(4) Silvio Peroni, Research Centre for Open Scholarly Metadata, Department of Classical Philology and Italian Studies, University of Bologna, Bologna, Italy and Digital Humanities Advanced Research Centre (/DH.arc), Department of Classical Philology and Italian Studies, University of Bologna, Bologna, Italy {silvio.peroni@unibo.it};
(5) David Shotton, Oxford e-Research Centre, University of Oxford, Oxford, United Kingdom {david.shotton@opencitations.net}.
Table of Links
- Abstract and Intro
- Related Works
- Methodology
- Data and services
- Discussion
- Conclusion, Acknowledgements, and References
Abstract
OpenCitations Meta is a new database that contains bibliographic metadata of scholarly publications involved in citations indexed by the OpenCitations infrastructure. It adheres to Open Science principles and provides data under a CC0 license for maximum reuse. The data can be accessed through a SPARQL endpoint, REST APIs, and dumps. OpenCitations Meta serves three important purposes. Firstly, it enables disambiguation of citations between publications described using different identifiers from various sources. For example, it can link publications identified by DOIs in Crossref and PMIDs in PubMed. Secondly, it assigns new globally persistent identifiers (PIDs), known as OpenCitations Meta Identifiers (OMIDs), to bibliographic resources without existing external persistent identifiers like DOIs. Lastly, by hosting the bibliographic metadata internally, OpenCitations Meta improves the speed of metadata retrieval for citing and cited documents. The database is populated through automated data curation, including deduplication, error correction, and metadata enrichment. The data is stored in RDF format following the OpenCitations Data Model, and changes and provenance information are tracked. OpenCitations Meta and its production. OpenCitations Meta currently incorporates data from Crossref, DataCite, and the NIH Open Citation Collection. In terms of semantic publishing datasets, it is currently the first in data volume.
Keywords— scholarly citations, bibliographic metadata, provenance, change-tracking, open science, OpenCitations
1. Introduction
OpenCitations is an independent not-for-profit infrastructure organisation for open scholarship dedicated to publishing open bibliographic and citation data using Semantic Web technologies. OpenCitations stores and manages information about scholarly citations, i.e. the conceptual links connecting a citing entity with a cited entity, in the OpenCitations Indexes. Hitherto, there have been four OpenCitations Indexes: COCI (https://opencitations.net/index/coci), the OpenCitations Index of Crossref open DOI-to-DOI Citations (Heibi et al., 2019b); POCI (https://opencitations.net/ index/poci), the OpenCitations Index of PubMed open PMID-to-PMID citations; DOCI (https://opencitations.net/index/doci), the OpenCitations Index of DataCite open DOI-to-DOI citations; and CROCI (https://opencitations.net/index/croci), the Crowdsourced Open Citations Index (Heibi et al., 2019a).
While the coverage of the OpenCitations Indexes has approached parity with that of commercial proprietary citation indexes (see https://opencitations.hypotheses.org/ 1420), there have been outstanding issues not formerly addressed by OpenCitations.
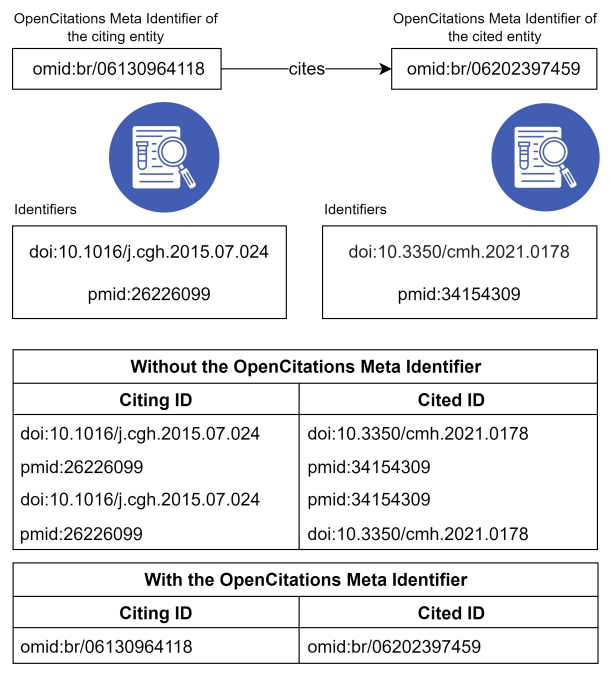
First is citation disambiguation. Sometimes, bibliographic resources will have been assigned multiple identifiers, such as a DOI and a PMID. In such cases, the same citation may be multiply represented in different ways depending on the data source. For example, OpenCitations will describe in COCI a citation between two publications using metadata derived from Crossref as a DOI-to-DOI citation, and in POCI the same citation using metadata derived from PubMed as a PMID-to-PMID citation. This duplication poses problems when counting the number of ingoing and outgoing citations of each document, a crucial statistic for libraries, journals, and Scientometrics studies. Use of OpenCitations Meta permits us to deduplicate such citations and solve the problems that such duplication would otherwise cause.
Second, the assignment of globally persistent identifiers to documents is not universal practice across all scholarly fields. Gorraiz et al. (2016) demonstrated that the Natural and Social Sciences communities adopt DOIs to a much greater extent than the Arts and Humanities community. From that research, carried out on Scopus and the Web of Science Core Collection, it emerged that almost 90% of the publications in the Sciences and Social Sciences are associated with a DOI, while in the Art and Humanities that figure is only 50%. In addition, concerning the Humanities, citations of ancient primary sources lacking DOIs (e.g. Aristotle) are required in many fields (e.g. in History). If a document has no identifier, its metadata does not respect the FAIR principles (Wilkinson et al., 2016) that scholarly digital research objects must be findable, accessible, interoperable and reusable. A globally unique and persistent identifier is critical to make metadata findable and accessible. Moreover, a bibliographic resource without an identifier prevents citations involving it from being described adhering to the FAIR principles. This is the reason why, according to the Open Citation Definition (Peroni & Shotton, 2018) governing the population of OpenCitations Indexes, any two entities linked by an indexed citation must both be identified by a persistent identifier coming from the same identifier scheme, for example both with DOIs, or both with PubMed IDs. For example, COCI (Heibi et al., 2019b) only stores citation information where the citing and cited entities are described in Crossref and both have DOIs. Citations involving publications lacking DOIs or other recognised PIDs have hitherto been excluded from the OpenCitations citation indexes.
But now, OpenCitations Meta solves the problems posed by bibliographic resources identified by multiple identifiers and also bibliographic resources that lack persistent identifiers, by associating a new globally persistent identifier to each document described in OpenCitations Meta - an OpenCitations Meta Identifier (OMID). In this way, all citations can be represented as OMID-to-OMID citations (Fig. 1). By providing a unique identifier for every entity stored in OpenCitations Meta, the entity’s OMID acts as a proxy between different external identifiers used for each entity, enabling disambiguation. Moreover, OpenCitations Meta can contain metadata for all scholarly publication, each identified by an OMID, without the mandatory need for an external persistent identifier to be provided by the source of the metadata.
Thus, thanks to OpenCitations Meta, metadata for all scholarly publications can now be stored by OpenCitations, and citations linking all such publications can be included within a new inclusive OpenCitations Index, of which the other indexes (COCI, DOCI, POCI, etc.) will be sub-indexes, according to the various input sources of the citation information.
Third is the previously poor temporal performance of the OpenCitations’ services, in particular API operations returning basic bibliographic metadata of citing and cited resources. This is because the OpenCitations Indexes themselves have hitherto contained only citation-related metadata (citations being treated as First Class data entities with their own metadata), but have not held bibliographic metadata relating to the citing and cited entities (title, authors, page numbers, etc.). Rather, those metadata have hitherto been retrieved on-the-fly by means of explicit API requests to external services such as Crossref, ORCID and DataCite
Over the past three years, to address the issues mentioned above, we have developed and tested the software we are now using to create a new bibliographic metadata collection, namely OpenCitations Meta, which we launched in December 2022. The software supporting this database is open source, and available at https://github.com/ opencitations/oc_meta. The metadata exposed by OpenCitations Meta includes the basic bibliographic metadata describing a scholarly bibliographic resource. In particular, it stores all known bibliographic resource identifiers for the bibliographic resource (e.g. DOI, PMID, ISSN, and ISBN), the title, type, publication date, pages, the venue of the resource, and the volume and issue numbers where the venue is a journal. In addition, OpenCitations Meta contains metadata regarding the main actors involved in the publication of each bibliographic resource, i.e. the names of the authors, editors, and publishers, each including their own persistent identifiers (e.g. ORCIDs) where available. It is our intention to add additional metadata fields (e.g. authors’ institutions and funding information) at a later date.
The process of generating OpenCitations Meta can be divided into two steps. The first step involves the curation of the input data. The curatorial procedure concerns the automatic correction of errors, the standardisation of the data format, and the deduplication of separate metadata entries for the same item. The deduplication process is based only on identifiers. This approach favours precision over recall: for instance, people are deduplicated only if they have an assigned ORCID, and never by other heuristics. After the normalization and deduplication stages, each entity is assigned an OpenCitations Meta Identifier (OMID), whether or not it already has an external persistent identifier (e.g. DOI, PubMed ID, ISBN).
The second step in populating OpenCitations Meta involves converting the raw input data into RDF (Linked Open Data format) compliant with the OpenCitations Data Model (OCDM) (Daquino et al., 2020), to enable querying such data via SPARQL. During this process, great attention is given to provenance and change-tracking: every time an entity is created, modified, deleted or merged, such changes are recorded in RDF, and are characterised by their dates of creation, primary sources, and responsible agents.
The rest of the paper is organised as follows. Section 2 reviews other semantic publishing datasets. Subsequently, in Section 3, the methodological approach adopted to produce OpenCitations Meta is presented in detail, starting with the curatorial phase (3.1), then describing error correction (3.2), moving to an explanation of the data translation to RDF according to the OCDM (3.3), and concluding with a description of the production of the RDF provenance and change-tracking data (3.4). Section 4 provides some descriptive statistics regarding the current OpenCitations Meta dataset. Finally, Section 5 discusses some present limitations of OpenCitations Meta, and a consideration of where OpenCitations Meta stands among similar scholarly datasets.
This paper is available on arxiv under CC 4.0 DEED license.
